The genome between stability and variability
PDF
The genome of living beings is permanently affected by anomalies, due to accidental errors or agents inside and outside the body. Fortunately, enzymes correct most of these anomalies and thus reinforce genetic stability. But they can also cause mutations that contribute to variability. Other systems can produce new genetic combinations. These mutations and recombinations are essential for the adaptation of populations to changes in the environment and therefore for evolution.
1. Introduction
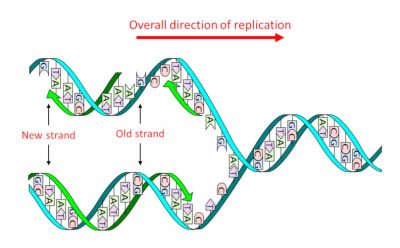
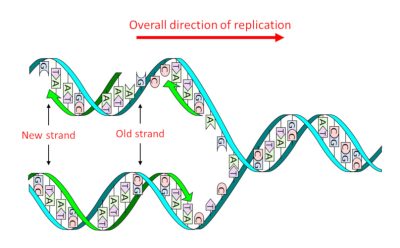
DNA consists of a double sequence of monomersmolecules which, by successive sequences with identical or different molecules, give rise to a polymer structure. (nucleotides) of which there are four types that only differ the nitrogenous base or nucleobase An organic nitrogen compound present in nucleic acids as a nucleotide in which it is bound to a ose, ribose in the case of RNA and deoxyribose in the case of DNA. In genetics, they are often simply referred to as the bases of nucleic acids : adenine (A), guanine (G), thymine (T) and cytosine (C). It is the precise sequence of these four types, and therefore the four nucleobases, that constitutes the “text” of the genetic message. Its structure in two complementary strands, where A is always facing T and G facing C, – the famous double helix (Figure 1) – has two major advantages:
- It allows, in a single step, thereplicationProcess to obtain two molecules identical to the initial molecule. into two identical daughter molecules (Figure 1), thus transmitting genetic information through cell divisions and sexual reproduction.
- It allows the repair of lesions that appeared on one of the strands as well as the creation of recombinant molecules, i.e. new genetic combinations. This double-stranded structure therefore promotes both stability and variability of genomes.
Genetic variations in DNA fall into two main categories: mutations and recombinations.
- Mutations are sudden changes in the genome of a living cell or virus. In multicellular organisms, if the mutations affect the germ cells intended to give gametes, they will be transmitted to the offspring and will therefore become inheritable.
- Recombinations consist in producing a new genetic combination from exchanges between existing genetic materials. There are several types of recombinations with very different biological mechanisms and roles. Classically, this definition only concerned DNA belonging to the same species. We now know that there are spontaneous genetic exchanges between different species, called transgenesis, that we will include in this category.
These two main types of variations are themselves divided into several modalities. We will endeavour to extract the elements most relevant to clarify their respective roles in population dynamics and evolution.
2. The mutations
2.1. General information
The stability of genetic messages encoded by DNA can be affected by errors of the enzyme that performs the replication, the replicase. This enzyme can thus put a nitrogenous base in place of another on the new strand. But apart from any replication, DNA is also permanently damaged. It is important to distinguish between lesion and mutation (see Genetic polymorphism & variation). Lesions are abnormalities in the physical structure of DNA that, in most cases, will prevent its replication. They cannot therefore be transmitted to the progeny. On the other hand, a mutated molecule has a normal physical structure. Only the sequence of the nucleobases – and therefore the information they contain – is modified. It can therefore replicate without problem and transmit the mutation to daughter cells.
The lesions are very diverse and there are quantities of repair enzymes, each highly specialized for a particular type of lesion. This is referred to as the “cell toolbox” for DNA repair (work awarded the Nobel Prize for Chemistry 2015). Paradoxically, it is these repair systems that, by mistake, will create the actual mutation from the primary lesion. These lesions can be caused either by agents internal to the body (or endogenous) or by agents from the external environment (or exogenous). Here are some examples to set the ideas.
- Endogenous agents: In warm-blooded animals, it is estimated that the DNA molecule can undergo 20,000 to 40,000 single-stranded cutsCuts in a bond between two adjacent nucleotides on a strand of a nucleic acid fragment. per cell and per day, as a result of molecular agitation. There can also be base losses, it is estimated that 10,000 T and C are lost and 500 A and G are lost, still per cell and per day. In these cases, we can even speak of spontaneous lesions. Among the most frequent endogenous agents are oxygen derivatives (free radicals or ROSA abbreviation for “Reactive oxygen species” or “reactive oxygen species”. Free radicals derived from oxygen, very reactive and very toxic. The abbreviation ROS is commonly used, even in French.), which are normal by-products of respiratory metabolism, they play an important role in oxidizing nucleobases that will need to be replaced. It would also be necessary to add the transposable elements,DNA sequence, sometimes called transposon, capable of moving autonomously in a genome, by a mechanism called transposition. These mobile DNA sequences are part of what are called dispersed repeats and are considered powerful drivers of evolution and biodiversity. we will discuss them further below.
- Exogenous agents: they can be physical (radiations) or chemical. The most common are ultraviolet (UV) rays, whose effect is generally limited to the skin because they are not very penetrating (see Cellular impact of solar UV rays). For example, it is estimated that sunbathing can cause, per cell and per hour, 60,000 to 80,000 abnormal chemical bonds between contiguous thymins of the same DNA strand, each of which being sufficient to block replication. If they are not all repaired, the cell dies: it is sunburn.
It is therefore clear that DNA stability is a dynamic process, resulting from a permanent balance between the production of lesions and their repair. These repair mechanisms do not operate at a constant level, they are subject to regulation.
A first type of regulation depends on the number of lesions in the cell. This phenomenon was first highlighted in the colibacillus (Escherichia coli), a bacterium that is one of the preferred subjects of study for geneticists. As early as 1974, it was assumed that there was a response, called SOS [2], which regulates the intervention of several repair systems according to the number of injuries. When there is a small number of lesions, this response increases the effectiveness of faithful repair mechanisms. But beyond a certain threshold of damage, these mechanisms are overwhelmed. The SOS response then induces the synthesis of a replicase able to cross specific lesions (see below), but with a certain error rate. This is called SOS mutagenesis. At the end of the 1990s, it was shown to facilitate the adaptation of the bacterial population to a hostile environment, at the cost of significant losses due to harmful mutations. It is a kind of last chance, hence the name SOS.

The second type of regulation concerns the fidelity of replication according to the type of organism. In all living cells, bacteria and others, this replication involves, in addition to DNA replication, a corrective enzyme system. Thanks to the old strand, this system corrects the errors made on the new strand by the replicase, which results in a very low error rate, in the order of 1 in 10 billion (10-10). Such accuracy is essential for organisms with large DNA molecules: 4 million base pairs in colibacillus and 3 billion in humans.
But in multicellular organisms, the number of cells can be very large: in the human species, it is estimated at one hundred thousand billion (1014). In these organisms, the majority of mutations are neutral, because the genome contains a large number of non-coding sequences (see Genetic polymorphism & variation). Even taking this into account, an error rate of 1 in 10 billion (10-10) is still too high. This is even more so since many mutations also occur outside replication. If there were only one copy of the gene per cell, many of them would carry deleterious mutations and the organism would not be viable. It is the interest, and the necessity, to have two chromosome sets for these organisms (diploidyProperty of a cell whose chromosomes it contains are present in pairs (2n chromosomes). The concept is generally to be contrasted with haploidy, a term referring to the ownership of cells with single copy chromosomes (n chromosomes). An organism or part of an organism is said to be diploid when its cells are themselves diploid.). Most mutations that inactivate one gene are recessive, so one functional copy is sufficient for the cell to function normally. In a nutshell, it can be said that diploidy acts as a spare wheel.
If we now look at the case of viruses, the situation is very different because most of which have very small genomes. For DNA viruses, there is generally no corrective activity associated with replicase, so the mutation rate is much higher, from 10-8 to 10-6 depending on viruses. This is even more true for viruses whose genome is composed of RNA, because RNA replicases are much less accurate than DNA replicases, the error rate is then 10-6 to 10-4.
However, in these RNA viruses, two categories must be distinguished according to the size of the genome. In those whose genome size is around 10,000 nucleotides, the fidelity of replication depends on the replicase alone. This is the case with influenza viruses (13,500 nucleotides) and HIV, which causes AIDS (10,000 nucleotides). In the latter, the rate of point mutations has been estimated at 1 in 10 per genome and per replication cycle. As the number of viral particles produced (virions) is of the order of 10,000 per day and per infected cell, it can be seen that the overall population in an infected host can exhibit considerable variability.
In RNA viruses with a genome of around 30,000 nucleotides or more, it is now known that there is corrective activity associated with the replicase. This is the case with Coronaviruses, including the SARS-CoV-2 responsible for the COVID-19 pandemic, whose genome contains 29,903 nucleotides. This corrective activity improves replication fidelity by a factor of 20, thus making it possible not to produce an excessive number of defective virions.
To put it more generally, in all viruses, whether DNA or RNA, there does seem to be a negative correlation between the rate of mutations and the size of the genome. For each viral species, a balance has been developed for the rate of mutations by natural selection. It should not be too high, so that the perpetuation of the viral species is not compromised, but still enough to create variability that allows the virus to bypass the defense mechanisms of its host. On this last point, the case of HIV is quite emblematic.
This leads us to make two remarks on the role of mutations in the evolutionary process:
- They play a key role in the adaptation of species to their environment because, by increasing genetic diversity, they are the material on which natural selection can act. What we have just seen with viruses is an example. The development of antibiotic-resistant bacteria, insecticide-treated insects, or herbicide-treated plants, are other examples.
- A genetic mutation is not inherently advantageous or disadvantageous, depending on the environment. For example, in the Kerguelen Islands, in the subantarctic, there is a wingless fly species, Calycopteryx moseley (Figure 3). This character, strongly disadvantageous in our regions, is on the contrary beneficial there because it prevents these flies from being carried into the ocean by the very strong winds that constantly sweep these islands.

Figure 3. Wingless fly, Calycopteryx moseley, living in the Kerguelen Islands, in an extremely windy environment. [Photos © Marine Pouvreau]
What are the different categories of mutations? They are classically distinguished by the size of the DNA segment concerned. In increasing order, these range from point mutations, where a single pair of nitrogenous bases is modified, to mutations that affect larger or smaller DNA segments, to tens of thousands of nucleotides. Not to mention the changes in the number of chromosomes. The last two categories mainly concern eukaryotic organisms, whose chromosomes are located in a particular compartment of the cell: the nucleus. These chromosomes each consist of a very long DNA molecule but in a complex “package”, composed of several families of proteins.
2.2. Point mutations
They can be due either to errors during replication or, more frequently, to poorly repaired lesions; they create variants of the same gene (alleles). In E. coli, it is the SOS system that causes the vast majority of mutations after UV irradiation. Most of the examples cited above relate mainly to point mutations.
2.3. Chromosome mutations
These are rearrangements produced by agents that cause DNA breaks, including radiations (other than UV). Several double-strand breaks can lead to more or less significant rearrangements depending on their number and the size of the segment concerned.
In the case of two very distant double-stranded breaks on the same chromosome, the entire segment between the breaks can be either (a) lost, causing the individual to die, (b) reversed at the same site (inversion), or (c) transferred to another chromosome (translocation) if other breaks have occurred on it. Inversions and translocations are quite frequent in natural populations. They interfere with the proper matching of chromosomes during meiosis The process of double cell division that takes place in the cells (diploids) of the germline to form gametes (haploids), or sex cells in eukaryotic organisms. and thus cause some sterility. As a result, they can be involved in speciationEvolutionary process that leads to the emergence of new living species that individualize from populations belonging to an original species. processes. Often, neighbouring species differ in chromosomal rearrangements.
2.4. Mutations of chromosome sets
This category has different mechanisms from the previous ones. These are no longer anomalies resulting from primary lesions but mechanical errors in cell division processes, mitosisrefers to the chromosomal events of cell division, the stage of the cell cycle of eukaryotic cells. This is the step of non-sexual/asexual duplication (unlike meiosis) of the chromosomes of the mother cell and their equal distribution in each of the two daughter cells. or meiosis. Some can lead to situations where individuals have more than two chromosome sets (2N), but always an integer number of sets (3N, 4N…), this is polyploidy (see Focus Polyploidy). They are viable because the “gene balance” is respected: all genes have the same number of copies. On the other hand, the 3Ns are sterile because the genetic balance of the gametes is necessarily abnormal. Meiosis obviously cannot evenly distribute an odd number of chromosomes among the gametes. The polyploids 4N, 6N or 8N are fertile. But they immediately create a new species, because any crossing with diploid parents would give sterile descendants, their meiosis being very unbalanced.
Other “failures” of meiosis can lead to a chromosomal imbalance in some gametes, which will therefore be found in the offspring. For example, they will be 2N-1 (monosomy) or 2N+1 (trisomy), we then speak of aneuploidy, which is a harmful situation because the genetic balance is no longer respected.
3. Recombinations
There are many recombination modalities with very different biological roles.
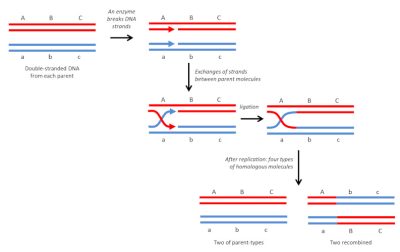
3.1. The homologous recombination
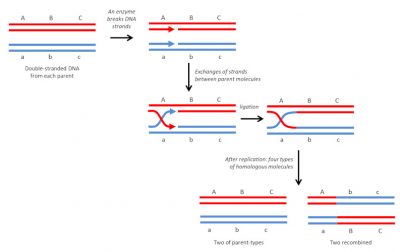
It is the most common and also the oldest known type. As its name suggests, it is done between identical DNA molecules (Figure 4). In diploid organisms, it occurs regularly, during meiosis, between homologous chromosomes, by crossover. It is a “soft” variability: the re-collection is done at exactly the same point, so there will be no mutations or modifications in the arrangement of genes on the chromosomes. This results in new and viable genetic combinations that may, in a given environment, have different adaptive capacities than parental combinations. In bacteria, it can also occur during conjugation [6] which allows genetic exchanges between cells [7].
This recombination mechanism is also involved in DNA repair. In diploid organisms, double-strand breaks caused on a chromosome by irradiation can be repaired by the presence of an intact homologous molecule in the segment concerned.
3.2. Other types of recombinations
To simplify, we will group together types of events with very different biological roles. They could be grouped under the term additive recombination because they result, in most cases, in the addition of DNA segments in genomes.
Among the most common mechanisms is the transposition of mobile genetic elements (called transposable elements) [8], which contain only a few hundred or thousands of nucleotides and possess, at a minimum, functions necessary for their “jumps” into the host genome. They occur in both bacteria and the most complex eukaryotes, sometimes in large numbers. In the former, they can be exchanged between different species during conjugation [6]. As they generally carry antibiotic resistance genes, they are one of the main factors in the very rapid spread of these resistances in pathogenic bacteria, with the serious medical problems that result.
In the human species, while our genes themselves, encoding all our proteins, constitute only about 2 to 3% of the DNA of the nucleus, the various families of transposable elements constitute nearly 50%. Most of them have been established in our lineage since very ancient geological times, long before the appearance of our species. The retrovirusFamily of RNA viruses with high genetic variability. Have an enzyme, reverse transcriptase, which allows the transcription of viral RNA into a particular DNA molecule capable of integrating with the DNA of the host cell., (to which HIV belongs), which are both virus and transposable element, alone represents 8% of our DNA. Fortunately for genome stability, only a small number of these elements are still mobile. As with “classical” mutations, the insertion of a transposable element can be harmful if it is done in a gene, but it can also bring interesting genetic innovations, either by modifying gene regulations or by their own functions. Multiple data show that they have contributed to the evolution of genomes and therefore species [9].
Finally, it is necessary to mention the phenomena of transgenesis, i.e. the genetic exchanges between different species. Since the late 1990s, it appears that they are much more frequent than we thought. The species barrier, which had been thought to be impassable under natural conditions, is actually quite porous, at least on an evolutionary scale. Again, this is a source of innovation that contributes to the plasticity of genomes and evolution.
4. Conclusion
We presented a brief overview of the different types of genome variations. We could not detail the enzyme systems involved in the repairs and recombinations, they alone would have required a complete article (if not two!). Readers interested in these mechanisms can refer to the following sites [10]. To conclude, two important points should be highlighted:
- Genetic variations occur randomly. They are not directed by the environment for adaptive purposes, as postulated in the Lamarckian vision of evolution. This has been abundantly demonstrated experimentally (Read Lamarck and Darwin: two divergent visions of the living world). However, contrary to what was believed until the 1980s, their frequencies can be modulated by the environment. We have seen this illustrated in bacteria with the increase in the rate of mutations by the SOS response and, in fruit fly and thale cress, with the stimulation of homologous recombination following irradiation. These are not the only factor capable of modulating the frequency of variations, many physiological stresses can lead to this result. For example, the application of antibiotics to bacterial cultures also triggers SOS mutagenesis and thus increases the frequency of mutations [11,12]. This subject is also covered in the article Adaptation: Responding to environmental challenges.
- This article is at the heart of what can be called the dialectic stability/variability of living beings. Two a priori antagonistic properties but in fact quite complementary. Both are essential for the survival of populations and the evolution of species.
Stability to allow a population to adapt sustainably to its environment when it is relatively stable and variability to facilitate genetic changes when the environment changes, allowing natural selection to work. The SOS response in bacteria is a good illustration of this since it is capable of performing either of these two functions, depending on environmental conditions.
References and notes
Cover image. [Source: © vitstudio; Image 134698571 via Shutterstock]
[1] http://atlasgeneticsoncology.org/Educ/DNAID30001FS.html
[2] R. Devoret (1993) Mécanisme de la mutagenèse SOS, Med/Sci vol.3, n°9, I-VII. (in french)
[3] J. Ducau et al (2000) Mutation Research 460:69-80
[4] J. Molinier et al (2006) Nature 442:1046-1049
[5] B.E. Rodgers & K.M. Holmes (2008) Dose response 6:209-221
[6] http://www.perrin33.com/microbiologie/genetique/conjugaison_3.php
[7] Video of Miroslav Radman: https://www.reseau-canope.fr/corpus/video/la-recombinaison-genetique-129.html
[8] D. Anxolabéhère, D. Nouaud & W.J. Miller (2000) Transposable elements and genetic novelties in eukaryotes. Med/Sci, I No. 11, vol. 16, I-IX.
[9] see “The surprises of evolution: viruses among our ancestors? “in www.lespiedsdansleplat.me
[10] http://www.cours-pharmacie.com/biologie-moleculaire/reparation-de-ladn.html or http://gec.sdv.univ-paris-diderot.fr/genetique/chapitre9.html
[11] S. Da Re & M.-C. Ploy (2012) Antibiotics and bacterial SOS response. Med Sci (Paris) 28:179-184 (in french)
[12] J. Blázquez, J. Rodrı́guez-Beltrán & I. Matic (2018) Antibiotic-Induced Genetic Variation: How It Arises and How It Can Be Prevented. Annu. Microbiol Rev. 72:209-30
The Encyclopedia of the Environment by the Association des Encyclopédies de l'Environnement et de l'Énergie (www.a3e.fr), contractually linked to the University of Grenoble Alpes and Grenoble INP, and sponsored by the French Academy of Sciences.
To cite this article: BREGLIANO Jean-Claude (May 30, 2021), The genome between stability and variability, Encyclopedia of the Environment, Accessed December 26, 2024 [online ISSN 2555-0950] url : https://www.encyclopedie-environnement.org/en/life/genome-between-stability-and-variability/.
The articles in the Encyclopedia of the Environment are made available under the terms of the Creative Commons BY-NC-SA license, which authorizes reproduction subject to: citing the source, not making commercial use of them, sharing identical initial conditions, reproducing at each reuse or distribution the mention of this Creative Commons BY-NC-SA license.