用DNA条形码描述生物多样性

新兴的高通量分子生物学技术使得我们可以鉴定环境中的生物物种。通过检测存在于环境中的DNA短片段(如从水、土壤、排泄物样本中),我们可以详尽地列出生态系统中的生物多样性的清单。这样的生物多样性调查可用来侦测环境中离散的物种,重构古代环境或者食物网等。这样的工作还为DNA溯源以及食品、化妆品的可靠性控制提供了前景。
1.通用的诊断工具
物种的鉴定从来都不是一个新话题,它关系到为各种目的(如食品、医药、文化等)从环境中获取生物的人类社会。如今,从物种水平来描述生物多样性已成为一个重大的科学和社会挑战。事实上,物种之间通过交互作用影响生态系统的功能、动态和演进,而为了理解这种作用,物种鉴定是一个必不可少的先决条件。在现今气候、土地利用、城市化等全球变化的大背景下,物种鉴定对保护、恢复生物多样性显得尤为必要。
过去物种的描述主要基于形态学标准,现在仍是如此,但目前越来越多地以分子标准,特别是DNA特征为标准(参见:“什么是生物多样性”。而DNA特征在研究难以获取形态特征的群体(比如微生物)、或者形态特征差别不大的群体(如线虫)时,就显得尤为重要了。
“生命条形码”[1]计划始于21世纪初,旨在提供一种多样性诊断工具,能够广泛应用于生态学、农学、海关管制等多个领域。这一举措有助于加快生物多样性的描述。这项工作立足于DNA条形码的概念,通过比较从某个体获得的标准化基因数据和馆藏标准和分类学家(分类学家的目的是描述现存的生命体并将它们分组到不同的分类单元,以便通过二分法等来鉴定、分类及辨识它们)鉴定的标准化基因数据,从而快速可靠地鉴定物种。
为此,采用的DNA条形码必须与标准的DNA片段相一致,其序列具有物种特异性。譬如,“生命条形码”计划的发起者们已经将一段编码细胞色素氧化酶Ⅰ(细胞呼吸链酶复合物亚基),缩写为COX1的线粒体基因的一个区域鉴定了出来。与大多数由细胞核基因编码的细胞色素氧化酶亚基不同,COX1这个亚基是由线粒体基因组编码的。除了刺胞动物,利用COX1的序列可将庞杂的动物种类加以区分的线粒体基因区域,作为鉴定不同动物物种的参考片段。
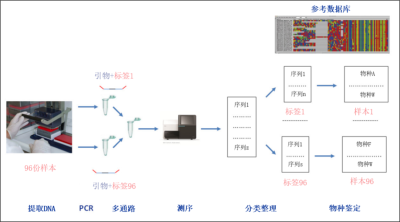
首先,从样本中提取DNA。随后,用已知的含有DNA标签的引物,对匹配上DNA条形码的目的区间进行PCR扩增。这样一来,从同一样本分离出的所有DNA片段都带上了特定的标签。然后将所有样本混合(多通路),进行高通量测序。测序完成后,获得的DNA序列可再通过它们的标签回归到原来的标本源。最后,比对获得DNA序列和参考数据库,即可鉴定样本中的物种。
DNA条形码的概念不仅被分类学家所利用,也被生态学家使用,他们广泛地使用DNA片段鉴定样本。即使生物体难以获取,我们也能通过这种策略来鉴定环境中的物种。毫无疑问,不仅微生物适用于这种方法;而且像动植物这种存在的宏观生物,也能凭借溯源它们遗留的DNA痕迹(如尸体、粘液、排泄物等)来检测。其原理是从环境样本(水、土壤等)中提取的DNA,然后通过PCR(聚合酶链式反应的缩写,英文缩写已成为常见的说法。PCR技术是一种体外的靶向复制技术,可以从组分复杂、目标含量稀少的样本中复制,得到大量确定长度的特异性双链DNA片段。每次PCR循环分为三步:加热使得DNA变性解为双链;引物在目的片段的两端杂交;DNA聚合酶作用下片段延伸。)从而扩增了目的DNA片段[2]。PCR引物(PCR反应中用到的寡核苷酸序列,通过对其限定可以特异性地扩增想要的序列。)可以特异性地甄别目标物种,例如像牛蛙这样的入侵物种,即使密度极低,也能通过池塘的水样检测到。反过来,也可对引物进行定义,这样就可以同时研究更大的物种范围。这种方法也被称为“元条形码”。这样的话,由PCR扩增的DNA片段就需要进行测序并和参考数据库相比对,以便将他们归属到不同的已知物种去(图1)。
2.用于生态学的分子生物学工具
要用元编码手段基于环境中DNA样本来有效鉴定物种,就必须定义方便的DNA条形码。这样的条形码必须在不同物种之间具有序列的差异,而在同一物种内具有一致性的序列,这样才能具备较高的鉴定能力(分辨率)。同时,这段序列必须由两段在不同物种间高度保守的区域所包围,以便能够在尽可能多的物种中同时扩增目标片段,换言之,它将拥有最大的分类学范围。另外,扩增的片段必须较短,以表征降解的基质。事实上,DNA在环境基质中会发生降解,因此很难扩增大于150核苷酸长度的片段。此外,由于样本的DNA数量通常很有限,而细胞内线粒体或者叶绿体的DNA片段拷贝数是细胞核DNA拷贝数的100到1000倍,因此最好采用线粒体或者叶绿体的DNA片段。同时,利用系统进化信息的DNA条形码(其差异性水平反映了物种间的差异性)可将未知物种与已知相关物种联系起来,因此大有裨益。利用专用的生物信息学工具从庞大的DNA序列数据库挖掘数据,可以为研究一组生物体定义最具相关性的条形码。这些生物信息学手段也可以用来评估条形码的分辨率和分类学范围。

收集沉积物样本(图片来源:© E·维勒斯列夫(E. Willerslev))和由宏条形码鉴定的主要物种(图片来源:© 约瑟夫·傅立叶(Joseph Fourier)阿尔卑斯站)。
一旦确定了DNA条形码,我们将按照以下的步骤来鉴定存在于某一环境样本中的所有物种(图1)。
采样过程必须严格遵从规范以避免污染(图2)。
对于不同的类型的研究样本(水、土壤、排泄物等)有相对应的采样方案,每份样本的DNA提取必须按照相应的提取方案开展。
提取到的DNA随即通过靶向到条形码区域的引物进行PCR扩增。在每段引物的一个末端都附上了一段对不同样本特异的短DNA标记,以便携带将不同序列分配到样本源的必要信息(图1)。这一阶段,可利用一种可特异性结合到特定物种DNA上的寡核苷酸,来遏制这一特定物种的扩增反应。举例来说,在利用捕食者的粪便来对其进行捕食习性分析时,可用此方法来阻遏捕食者自身DNA的扩增,因为其自身DNA大量存在,可能掩盖我们对某些猎物信息的检测。
PCR结束后,每份样本都用扩增得到的相应物种DNA条形码的混合物(扩增子)表示。
在多种方法联用的步骤里,不同样本来源的复制子要混合起来(图1)。
接着对扩增子进行高通量测序。以前在实践中无法想象的元条形码方法,通过下一代测序技术得以蓬勃发展。目前的技术已经能够对数百万个DNA分子进行多通路并行测序[3]。从几百份样本开始,每份能够获得数千个序列,这对于提供相关信息来说绰绰有余。
测序完成后,基于引物标签将序列按照样本源信息分类整理,随后通过比较样本序列和参考序列,即可将它们归属到物种水平了。
整套流程下来,生物信息学工具起到了不可替代的作用:整理数据,建立参考数据库,借由这些数据库将序列归属到不同的分类,确立标签的列表以及处理测序错误等。
在不鉴定物种的情况下,所获的DNA序列可用来确立操作单元(MOTUS,即分子操作分类单元),并据以此来量化样本中的生物多样性。在处理那些未被描述过的、甚至是无法培养的微生物时,通常就可以用这种方法。
3.元条形码方法用于描述环境样本
迄今为止,生物多样性的描述方法一直采用较为繁琐的方法,而元条形码方法提供了一条新路。这种方法为研究陆地和水生生态系统的功能和演变开拓了新思路,当然这需要我们提前掌握有关物种群落及其内部互作的知识[4]。相比较而言,使用元条形码方法来描述土壤样本的物种多样性更为有效,尤其是在生物个体难从形态上加以鉴定之时。事实上许多大型土壤动物的情况正是如此,它们在生态系统中发挥着不可或缺的作用,例如蚯蚓、昆虫、弹尾虫等。元条形码还能取代传统的植物学研究,尤其是在像热带雨林这样生物多样性极度丰富的环境中。亚马逊流域包含11,000种树木,其中的一半濒临灭绝。传统的植物学研究手段会导致多达20%的属被忽略,而采用DNA条形码技术将获得更佳的分辨率。
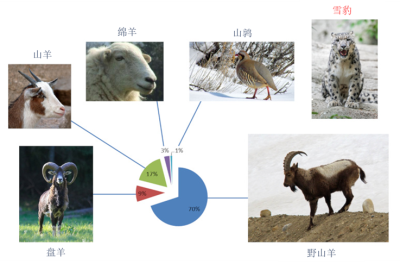
熟悉濒危动物食物习性对实行保护措施至关重要。在研究行为谨慎或生存于人迹罕至之地的稀有物种时,其猎物信息尤难以辨识。雪豹就是这样一种典型的濒危物种,它们是当地家畜的主要捕食者,因而在中亚地区很不受欢迎,这导致雪豹被驱逐,本来当时可以实施临时措施。事实上,谢赫扎德(Shehzad)和他的合作者已经借助于宏条形码技术,分析了从蒙古南部获得的雪豹粪样,成功地发现雪豹的食物谱主要由野生有蹄类动物构成。(图片来源:盘羊—Wikicommons;山羊—©安德烈·卡沃斯·阿卡(André Karwath Aka)(CC BY-SA 2.5) via Wikimedia Commons;绵羊—©沃蒂高顿(Vertigoten)(CC BY-SA 2.0) via Wikimedia Commons;山鹑—©伊万奇(evancj via Wikimedia Commons);雪豹—©埃里克·基尔比(Eric Kilby)(CC BY-SA 2.0) via Wikimadia Commons;野山羊—©苏亚旺什(Ksuryawanshi)(CC BY-SA 4.0) via Wikimedia Commons)
元条形码同样为传统的根据粪便或胃的内容物来进行食性评估的方法提供了新手段(图3)[5]。通过视觉的片段辨认食草动物体内的植物角质层(在植物体中覆盖气管的保护层,尤在叶中。角质层由连续的蜡质沉积而成,覆盖一层疏水脂肪酸,亦即角质)碎块,食草动物抑或是食肉动物体内的猎物残留角质层,只能提供的片面的信息。因此对被捕食者的DNA特征分析,将构建起一种破译生态系统中营养关系网(关于器官、组织营养)的高效手段。
最后,元条形码技术在重构古生态系统方面效果尤为显著,即使参与构成这些生态系统的物种早已消失。传统的研究手段,比如对大型化石和花粉的研究,实施起来十分繁杂,并且分辨率极低。永久冻土层(地质学术语,指的是温度连续两年以上保持在0℃以下的土层。永久冻土层占地球表面面积的20%以上。英文用“permafrost”表述,俄语用“pergelisol”表示。永冻层被一层叫做“活土层”的土壤所覆盖,夏季来临时解冻,植物得以生长。全球变暖效应使得永冻层逐步解冻,给环境造成了巨大影响:甲烷释放、病原微生物释放等)(参见:永久冻土)2万多年前的样本比花粉分析样本的分辨率要高得多。西伯利亚柏胶醇(指代西伯利亚的永久冻土的术语)不同土层的分析表明,前冰期的草原由非禾本草本植物构成,而在后冰期被禾本植物所取代。与此同时,对在冰期后消失的猛犸象的胃内容物化石的分析表明,它们主要以采食这些非禾本草本植物为生。这样一来,元条形码使我们能够将生态系统中已灭绝动物的食性同发生剧变的植物多样性联系起来。
由于新测序技术的蓬勃发展,元条形码技术已经替代传统手段用于描述环境样本生物多样性[6]。除此以外,通过对不同条形码的综合分析,该方法还为对同一样本进行综合研究提供了新思路。我们可以想象,对于肠道或者瘤胃寄宿的微生物群(指生活在在宿主(动、植物)体内某一特定环境(微生物组防治微生物感染的生态学方法)的所有微生物(细菌、酵母、真菌、病毒)的合集,重要的一例便是生活在肠道中微生物,以前称为“肠道菌群”。),寄生虫的生活周期以及某一物种的食性,都可以通过它的粪便,进行综合分析。这项技术的应用范围不局限在生态学研究,还扩展到通过DNA特征分析其复杂基质的组成分析的领域中,比如法医、农产品和海关管制等。
参考资料和说明
封面照片来源:© 雅克·卓艾雅德
[1] http://www.barcodeoflife.org
[2] http://www.ens-lyon.fr/RELIE/PCR/principe/principe.htm
[3] https://www.ebi.ac.uk/training/online/course/ebi-next-generation-sequencing-practical-course/what-you-will-learn/what-next-generation-dna-
[4] Pompanon F, Coissac E, Taberlet P (2011) Metabarcoding, une nouvelle façon d’analyser la biodiversité. Biofutur, 319:30-32.
[5] Shehzad W et al (2012) Prey preference of snow leopard (Panthera uncia) in South Gobi, Mongolia. PLoS ONE 7(2): e32104. doi:10.1371/journal.pone.0032104
[6] Joly D, Faure D & Salamitou S (2015) Empreinte du vivant, l’ADN de l’environnement. Le Cherche Midi, 192 p.
环境百科全书由环境和能源百科全书协会出版 (www.a3e.fr),该协会与格勒诺布尔阿尔卑斯大学和格勒诺布尔INP有合同关系,并由法国科学院赞助。
引用这篇文章: POMPANON François, SHEHZAD Wasim (2024年3月12日), 用DNA条形码描述生物多样性, 环境百科全书,咨询于 2025年4月7日 [在线ISSN 2555-0950]网址: https://www.encyclopedie-environnement.org/zh/vivant-zh/dna-barcodes-to-characterize-biodiversity/.
环境百科全书中的文章是根据知识共享BY-NC-SA许可条款提供的,该许可授权复制的条件是:引用来源,不作商业使用,共享相同的初始条件,并且在每次重复使用或分发时复制知识共享BY-NC-SA许可声明。









