共生与进化:真核细胞的起源

真核生物(动物、植物、真菌)的细胞与原核生物(古菌和细菌)的细胞区别在于是否存在特异性的细胞器,如:细胞核(遗传信息储存的场所)、线粒体(细胞呼吸的场所)或叶绿体(植物光合作用的场所)。线粒体和叶绿体中存在DNA,具有独特的生物化学和结构特征,因此人们认为它们是古菌通过内共生作用逐渐融入宿主细胞中形成的。有一种可能成立的假设,即如今的真核生物起源于一个意外俘获了变形菌的古老祖先,而这个变形菌正是如今的线粒体。如果这一假设成立,还可能存在一些细胞意外融合了蓝细菌(蓝藻),进而形成叶绿体。同时,它们得到了进行光合作用的能力,实现了自养代谢——一项植物的特性。在整个过程中,共生体间的基因转移、细胞核对某些细胞器蛋白质的编码以及基因产物转移到细胞中,使得这些原核生物与宿主细胞紧密结合。因此,内共生现象是进化过程中真核生物多样性出现的主要原因。由此,光合作用得以在多种生物中进行:红藻和绿藻、通过初级内共生形成的绿色植物、褐藻和许多通过二次甚至三次内共生形成的其他生物。
1.真核细胞是嵌合体
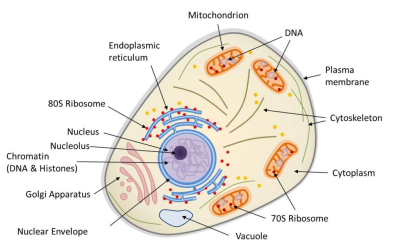
动物细胞是区室化的,由内膜系统(核膜、高尔基体、内质网、液泡等)、线粒体(受双层膜限制)和胞质内的细胞骨架组成。细胞核和线粒体都含有DNA。核糖体(蛋白质合成机器)以两种形式存在:70s核糖体(存在于线粒体中)和80s核糖体(通常与网状结构相关联)。(图1 Mitochondria 线粒体;Endoplasmic reticulum 内质网;80S Ribosome 80S核糖体;Nucleus 细胞核;Nucleolus 核仁;Chromatin 染色质;Histones 组蛋白;Golgi Apparatus 高尔基体;Nuclear Envelope 核膜;Vacuole 液泡;70S Ribosome 70S核糖体;Cytoplasm 细胞质;Cytoskeleton 细胞骨架;Plasma membrane 质膜)
真核生物(单细胞或多细胞生物,其细胞拥有由膜界定的细胞核和细胞器(内质网、高尔基体、各种质体、线粒体等)。真核生物与细菌和古菌构成了生物的三种类型)包括多细胞生物(动物、植物、真菌)和单细胞生物(如原生动物)。真核细胞(图1)的主要特征是存在细胞核(在原核生物中,基因组很少被膜包围)、细胞质中含有许多细胞器,如线粒体【核细胞(植物、藻类、动物)胞质器:作为细胞呼吸的场所,线粒体通过“克雷布斯循环”将消化产生的有机分子(葡萄糖)的能量转化为细胞直接使用的能量(ATP)。这一反应需要氧气的存在并释放CO2,因此在碳循环中发挥着重要作用。线粒体起源于20亿年前融入真核原细胞的原核生物(α-变形菌)。】(存在于所有真核细胞中,负责呼吸作用)和叶绿体【进行光合作用的真核细胞(植物、藻类)的胞质器:叶绿体作为光合作用场所,产生氧气(O2)并在碳循环中发挥着至关重要的作用:它们利用光能固定CO2并合成有机物质。因此,它们负责植物的自养过程。叶绿体源自大约15亿年前,光合型原核生物(蓝细菌型)在真核细胞内发生的内共生。】(存在于广义的植物,如陆生植物和藻类,负责光合作用【光合作用是一种生物能量过程,允许植物、藻类和一些细菌利用阳光从大气中的CO2合成有机物质。太阳能被用于氧化水和还原二氧化碳,从而合成有机物质(碳水化合物)。水的氧化导致大气中氧气(O2)的形成。光合作用是自养的基础,它是叶绿体在细胞内协同运作的结果】。这些细胞器经常被细胞骨架移动或重组(图1)。
真核细胞的细胞核被核膜(双层膜)包被(图1)。它包含真核细胞特有的核基因组,即在DNA(脱氧核糖核酸)中编码的个体遗传物质。当我们提到真核生物的基因组时,我们通常指的是这个基因组。然而,真核细胞在细胞器内也含有非核基因组:
-线粒体基质内的线粒体基因组(图1);
-叶绿体基质内的叶绿体基因组(例如植物或藻类)。
这三个基因组的DNA的组织方式并不相同。在细胞核中,基因组分布在几个DNA分子上,线性排列成分化良好的染色体。DNA包含所有编码序列(能转录成信使RNA(mRNA)并翻译为蛋白质)和非编码序列(不转录,或转录为RNA但不翻译)。细胞核基因组的三维构型具有重要的功能意义:DNA缠绕在组蛋白周围再经过多次盘曲折叠(或称“缩合”),使得大量的遗传信息被包装到微小细胞核中。线粒体或叶绿体DNA的组织方式则完全不同:它们通常是环形的,很少是线性的(植物线粒体),通常没有内含子,与组蛋白没有联系。
原核生物【一种微生物(通常为单细胞),具有简单的细胞结构,没有细胞核,几乎都没有内部区室(唯一的例外是蓝藻细菌中的类囊体)。生物三大类型中的两个是原核生物:古菌和细菌。原核细胞(细菌和古菌)是一类单细胞原核生物,主要存在于极端环境中(厌氧、高盐度、极热等)。卡尔·沃斯和乔治·E·福克斯(1977年)的系统发育研究将古菌与其他原核生物(细菌)区分开来。目前,生物被认为包括三个群体:古菌、细菌和真核生物。)】没有细胞核,DNA是环状的(或在极少数情况下呈线性),组织结构类似于叶绿体或线粒体。因此,DNA的复制、转录和翻译直接在细胞质中进行。然而,必须强调的是,古菌只是表面上与细菌相似:它们的新陈代谢有很大差异,参与复制、转录和翻译过程的机制和蛋白质表现出与真核生物相似的特征。最后,原核生物一般是没有内部区室的,即使存在,也没有那么复杂(蓝藻是一个例外)。最重要的是,当区室存在时,它们在细胞中是不可移动的:我们最初发现的细胞骨架并不会移动细胞成分。
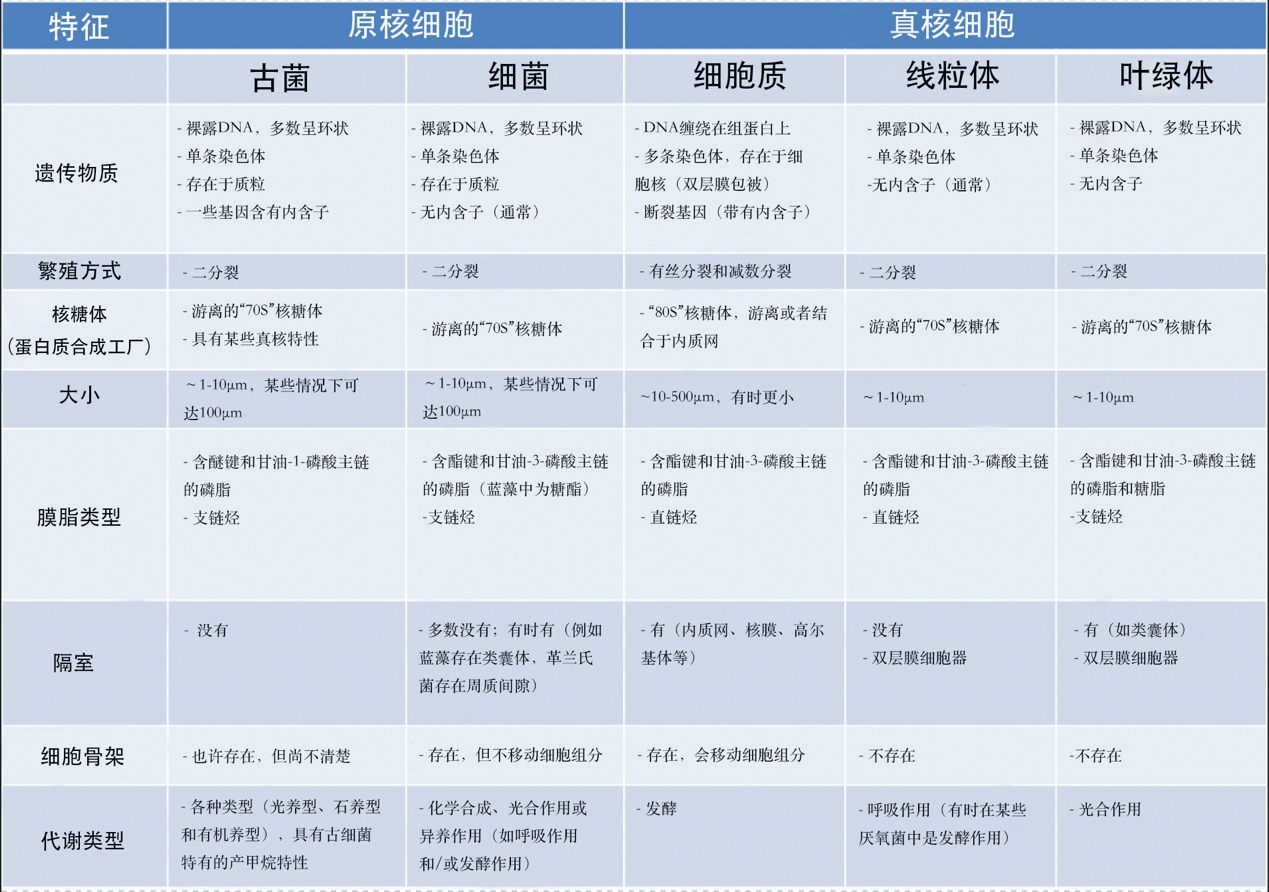
表1比较了原核细胞和真核细胞(及其线粒体、叶绿体)的性质。说明线粒体和叶绿体具有许多与原核细胞相同的特征。除DNA结构之外,真核细胞的细胞器是由先前存在的细胞器形成的,通过分裂繁殖,就像细菌一样。同样,它们有相同的蛋白质合成机制(基质或基质中的游离70S核糖体【核糖体是由RNA和核糖体蛋白组成的庞大复合体,它使mRNA翻译成蛋白质。核糖体是所有细胞(原核生物和真核生物)共有的,但根据生物体的不同而有所变化:真核生物中为80S核糖体,原核生物和细胞器(线粒体、叶绿体)中为70S核糖体。】),而在真核细胞的细胞质中,这种机制由80S核糖体组成,有时固定在内质网膜上【内质网是真核细胞胞质中的膜网络,对细胞新陈代谢(脂质和蛋白质合成、钙储存)至关重要。与核糖体结合,是细胞内合成分泌到细胞外的蛋白质以及构成细胞器膜的蛋白质和脂质(高尔基体、溶酶体、线粒体、细胞核、核糖体、囊泡等)的地方。】。最后,细菌也有线粒体的代谢作用(即呼吸作用),在某些特殊情况下,还有叶绿体的代谢作用(即光合作用)。另一方面,真核细胞的特点是存在一个活性蛋白质网络,即细胞骨架,一个能够移动的自组织系统,负责定位和移动细胞中的细胞器。在原核生物中,这种蛋白质网络是静态的,甚至不存在的,在线粒体和叶绿体中也几乎没有这种骨架。
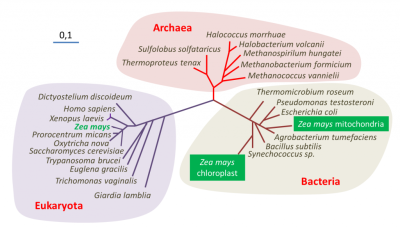
表明了玉米(Zea mays)中包含的三个基因组(细胞核、线粒体和叶绿体)的位置——聚球藻(Synechococcus)是一种蓝藻[来源:改编自参考文献[1](图2 Archaea 古菌;Bacteria 细菌;Eukaryote 真核生物;Halococcus morrhuae 嗜盐球菌属 嗜盐球菌;Halobacterium volcanii 嗜盐杆菌属 火山盐杆菌;Methanospirillum hungatei 甲烷螺菌属 亨氏甲烷螺菌;Methanobacterium formicium 甲烷杆菌属 甲酸甲烷杆菌;Methanococcus vannielii 甲烷球菌属 万氏甲烷球菌;Sulfolobus solfataricus 硫化叶菌属 硫磺矿硫化叶菌;Thermoproteus tenax 热变形菌属 附着热变形菌;Thermomicrobium roseum 热微菌属 玫瑰色热微菌;Pseudomonas testosterone 假单胞菌属 睾丸酮假单胞菌;Escherichia coli 埃希氏菌属 大肠杆菌;Zea mays mitochondria 玉米线粒体;Zea mays chloroplast 玉米叶绿体;Agrobacterium tumefaciens 农杆菌属 根癌农杆菌;Bacillus subtilis 芽孢杆菌属 枯草芽孢杆菌;Synechococcus sp. 蓝细菌聚球藻属 聚球藻;Dictyostelium discoideum 网柄菌属 盘基网柄菌;Homo sapiens 人属 智人;Xenopus laevis 爪蟾属 非洲爪蟾;Zea mays 玉米;Prorocentrum micans 原甲藻属 海洋原甲藻;Oxytricha nova 尖毛虫属 尖毛虫;Saccharomyces cerevisiae 酵母菌属 酿酒酵母;Trypanosoma brucei 锥虫属 布氏锥虫;Euglena gracilis 裸藻属 纤细裸藻;Trichomonas vaginalis 毛滴虫属 阴道毛滴虫;Giardia lamblia 贾第虫属 蓝氏贾第鞭毛虫)
利用DNA测序技术进行基因组序列分析提供了关于生物进化史的信息,特别是关于它们的亲缘关系,也称为系统发育【研究相关物种之间联系的学科称为“系统发育学”(phylogenetics)。它允许追踪生物从共同祖先演化的主要阶段,并建立生物之间的亲缘关系。】(参阅 什么是生物多样性?以及遗传还是趋同?)。对玉米核基因组及其线粒体或叶绿体基因组进行分子系统发育分析,使确定这种植物在生命树中的系统发育位置成为可能(图2)。分析表明,三个谱系(其中两个属于细菌)在结构和功能上纠缠在一起,被认为是一个单一的有机体——显示了该物种的三重起源。
所有这些特性表明,真核细胞是一种嵌合体,它既包含真核细胞的特征成分(细胞核),又包含具有典型原核特性的细胞器(叶绿体、线粒体)。
1925年,爱德华·查顿(Edouard Chatton)[2] (他命名了这两种细胞类型)提出了原核细胞和真核细胞之间的区别,然而这两种细胞类型的说法直到20世纪50-60年代才被接受。自19-20世纪之交,人们观察到了真核细胞的嵌合性。1883年,法国植物学家安德烈亚斯·辛珀(Andreas Schimper)提出光合作用是不同生物体结合的结果,随后俄罗斯生物学家康斯坦丁·梅列什科夫斯基(Constantin Mereschkowsky)给出了确凿论据,即某些细胞来自两种不同类型细胞的结合(内共生)。梅列什科夫斯基(Mereschkowsky)在其1905年的文章[3]中提出了三个基本观点:(a)叶绿体是一种蓝细菌,在进化的早期就与异养宿主建立了共生关系,(b)宿主本身也是更大的异养宿主间早期共生的产物,一种较小的微球菌型内共生体形成了细胞核,以及(c)植物的自养能力完全从蓝细菌继承而来。梅列什科夫斯基没有考虑线粒体的起源。这要归功于法国微生物学家保罗·波蒂埃(Paul Portier),他在1918年的一篇文章[4]中写道:“所有生物,包括所有动植物,都是由两种不同生物结合构成的。每一个活细胞都包含一种特殊结构,细胞学家称之为“线粒体”’。 对我来说,这些细胞器只不过是共生细菌,我称之为共生体。”这些观察结果没有引起科学家们的更多关注,这一理论因此失宠,尤其是因为质体和线粒体的培养实验(在19世纪,这一实验被认为是证明质体和线粒体是细菌起源的证据[5])失败了。1970年左右,美国微生物学家林恩·马古利斯运用电子显微镜、生物化学和分子生物学等新的细胞研究方法,提出了真核细胞细胞器内共生起源的理论。
2.真核细胞是如何演化的?
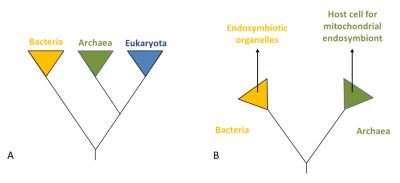
(A)一种“三界”系统,其中真核谱系和古菌谱系有相同的起源,两个谱系一样古老。(B)一种“二界”系统,来自最近的系统发育分析。
(图3 Bacteria 细菌;Archaea 古菌;Eukaryote 真核生物;Endosymbiotic organelles 内共生细胞器;Host cell for endosymbiont mitochondrial 内共生线粒体宿主细胞)
人们提出了多种假说来解释真核细胞的出现(约15亿年前,也就是在第一批原核生物出现近10亿年后)。这个问题可以用多种方式来解释,这取决于是否综合考虑了古生物学证据、能量方面、真核细胞特征的起源或不同原核和真核系之间的相互关系[6]。图3展示了真核生物和其他谱系(古菌和细菌)的起源。
一些模型认为,真核生物是由一个祖先谱系在进化期间经过连续突变产生的。另一些模型则认为,真核生物是由原核细胞的共生组合产生的,而这种融合导致了原核生物向真核生物过渡。这些不同的假说可以通过实验来部分验证,特别是通过对现有生物(原核或真核)基因组的分析[7,8]。
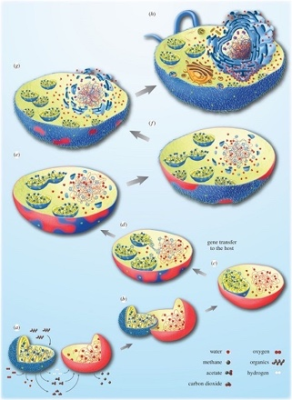
(a-h)描述从依赖氢气的古菌宿主细胞(红色)到兼性厌氧a-变形菌(蓝色),再转变为真核细胞的各种步骤。最初,这两种生物都生活在附近,古菌需要细菌产生的氢,而细菌无法承受其自身代谢产生的氢气积累。于是细菌的融合随之而来,这种转变伴随着两个生物体之间的基因转移(c),同时伴随着细胞核的建立(e-h)。[来源:经作者许可转载(见参考文献[5]) © 2015 (CC BY 4.0)](图4 water 水;methane 甲烷;acetate 醋酸酯;carbon dioxide 二氧化碳;oxygen 氧气;organics 有机物;hydrogen 氢;gene transfer to the host 基因向宿主转移)
在这种背景下,细菌和古菌之间密切互动,其互动形式与现今的许多共生关系相同。一般来说,这可能导致图4所示的情况,即细菌共生体将被原始宿主保留并最终驻留在宿主体内。在这种情况下,宿主并不是以共生体为食,因此融合不是由于吞噬现象造成的——尽管吞噬作用无疑增加了真核细胞进化过程中内共生体融合的频率 [5]。
真核细胞的起源也与细胞核有关,细胞核是真核细胞的标志性结构。线粒体形成后,宿主体内诞生了一种新的膜系统,即核膜,这可能是由于细菌脂质组成的膜囊聚集导致的。伴随着宿主和共生体间的基因转移,这种细胞核与细胞质的分离使得RNA剪接与DNA翻译过程分开。在这种情况下,选择压力可能导致了新形成的细胞核和细胞质之间区室化的固定[5](图4)。
因此,目前已知的所有真核生物都来自一个古老的祖先,它在前寒武纪时期融合了一种变形菌,即线粒体。这一步是至关重要的:这次融合与真核细胞的起源密不可分。原核细胞内的能量是匮乏的,这也是驱动这种细胞进化的一个主要因素:只有具有线粒体的细胞才有足够的能量实现真核细胞的复杂性,这就是为什么没有原核生物到真核生物的中间过渡态。通常认为,只有在线粒体融合完成之后,才有部分细胞获得了真核细胞的特征(细胞核、区室化),并且在某些特殊情况下,融合蓝细菌,这就是叶绿体的起源。同时,它们获得了进行光合作用的能力,因此能够自养代谢。
最近,一些研究揭示了线粒体共生的时间[10],他们是依据真核细胞核中存在的细菌基因的时间(即真核细胞核与游离细菌中发现的同源基因分离的日期)。他们发现,许多基因,包括一些导致真核细胞复杂性的基因,很可能是在线粒体形成之前获得的。这并不意味着线粒体进入的细胞像现在这样复杂,但它可能已经能够吞噬细胞。这种特性(真核生物特有的,依赖于细胞骨架的流动性)可能有助于线粒体的建立。因此,关于真核生物复杂性的演化仍然是推测性的,但可能在线粒体形成前就已经开始了。毫无疑问的是,线粒体形成进一步加速了这种复杂性的演化。
3.叶绿体的内共生起源
在白细胞或许多原生动物(图5)的吞噬过程中,被摄入的细胞通常被直接消化(如猎物),但有时它们会永久滞留在细胞中(内共生体)。因此,在内共生过程中,细胞器是通过吞噬作用内化而产生的,而不是在真核生物内消化原核生物的结果(图5)。陆生植物中的叶绿体是如此,与之相近的红藻和绿藻也是如此[11,12]。
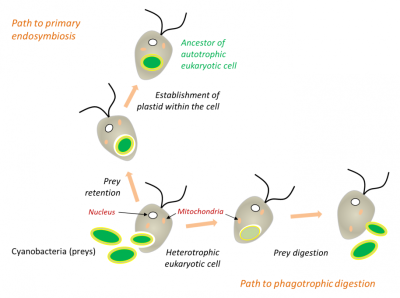
在吞噬过程中,被摄取的猎物通常被直接消化,但有时可能永久滞留在细胞中。细胞质膜包覆在原核生物周围,并将其隔离在内吞囊泡内。然后,当原核生物被融合到真核细胞中时,囊泡的膜消失了,位于蓝细菌两层膜之间的肽聚糖层也消失了[见参考文献 [9][10]。
(图5 Path to primary endosymbiosis 初级内共生途径;Path to phagotrophic digestion 吞噬消化途径;Heterotrophic eukaryotic cell 异养真核细胞;Cyanobacteria (preys) 蓝细菌(猎物);Nucleus 细胞核;Mitochondria 线粒体;Prey digestion 猎物消化;Prey retention 猎物滞留;Establishment of plastid within the cell 细胞内质体的形成;Ancestor of autotrophic eukaryotic cell 自养真核细胞的祖先)
初级和次级内共生
在进化过程中,内共生事件反复发生,导致特定生物的形成。在初级内共生过程中,真核细胞融合活的原核生物。因此,绿系植物(例如属于陆生植物的红藻和绿藻)的叶绿体源自含有蓝细菌的初级内共生体。在一些真核生物中,线粒体因适应厌氧环境而演化,但从未消失:它们产生特定的线粒体,如氢化酶体(hydrogenosomes),进行产氢发酵(例如某些纤毛虫)[14],但也产生小的细胞器,只参与宿主细胞的生物合成,如纺锤剩体(mitosomes)[15]。
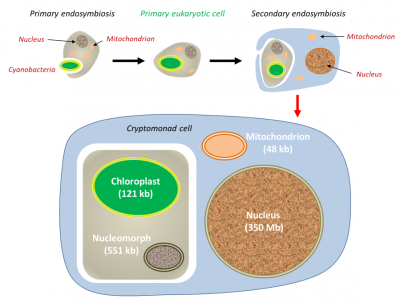
在这种情况下,红藻细胞核(初级宿主)以退化的细胞核(即核形体)的形式存在于次级宿主中,但基因组非常小(仅551 kb)。叶绿体和线粒体的基因组也大大减少。(图6 Primary endosymbiosis 初级内共生;Primary eukaryotic cell 初级真核细胞;Secondary endosymbiosis 次级内共生;Nucleus 细胞核;Cyanobacteria 蓝细菌;Mitochondrion 线粒体;Cryptomonad cell 隐滴虫细胞;Chloroplast 叶绿体;Nucleomorph 核形体)
4.原核生物与真核细胞的融合
所有这些品系都有一个共同的特点:内共生体存在强烈遗传退化。与大肠杆菌(Escherichia coli)等游离变形菌相比,线粒体丢失了99%的基因。在极端情况下,氢化酶体(hydrogenosomes)和纺锤剩体(mitosomes)根本没有基因组!与单细胞游离蓝细菌相比,绿系质体显示出95%的遗传退化:蓝细菌的基因数量从几千个降低到叶绿体中的100到200个左右……寄生植物大花草属(Rafflesia)中的质体甚至没有基因组。
这种退化显然是因为缺乏自由生活甚至是某些代谢功能所必需的基因。例如,与所有革兰氏菌【革兰氏菌是通过一种名为革兰氏染色的染色技术检测到的细菌:它们在显微镜下呈现粉红色。染色技术基于细菌的细胞膜和细胞壁的特征。然而,这并不是一个系统发育分类因素:事实上,“革兰氏阳性”组和“革兰氏阴性”组都是非单系的。革兰氏阳性和革兰氏阴性细菌细胞壁由一层肽聚糖构成,肽聚糖的主要结构可以分为两部分:多糖部分和肽部分。这层肽聚糖保持细胞的形状并保护细胞免受渗透压的影响】一样,蓝细菌的两层膜间有一层肽聚糖,这对于在低渗透压【渗透压指每升溶液中具有“渗透活性”的粒子的摩尔数。是一个与溶液中粒子施加的渗透压相关的概念,与渗透相关。蔗糖是一种小型的渗透活性分子,而淀粉是一个巨大的渗透不活性葡萄糖聚合物。在一个区室中蔗糖的积累导致该区室中的渗透压增加,而淀粉则不是这种情况。】的自然环境中维持细菌的结构至关重要。一旦原核生物融合到宿主细胞,原核生物将处于细胞质中,其渗透压非常接近原核生物自身内部介质的渗透压。显然,此时将不再需要肽聚糖层,于是叶绿体中有关肽聚糖层的基因逐渐丢失(灰胞藻除外)。
尽管细胞器基因组正在退化,但已知的细胞器蛋白质库(蛋白质组)仍然与游离细菌蛋白质组相似:因此,这些额外的蛋白质可能弥补了基因组的缺失。它们的编码实际上由宿主核基因组负责:细胞核基因在细胞质中翻译成蛋白质,通过转运肽(一种肽序列,位于细胞质中新合成蛋白质的NH2末端,定向到特定的细胞器(线粒体等)以执行其功能,也称之为定向肽)到达细胞器。这种基因产物的重新定位,对于原核生物融合到宿主细胞是必要的。负责转移的寻址机制是质体和线粒体的融合创新,是细胞内共生体的又一个新功能。这种机制允许细胞质中合成的蛋白质,通过跨膜转运进入线粒体和叶绿体中发挥作用。这种寻址涉及大量进化起源复杂的蛋白质:原核或者真核来源;由细胞器或者细胞核编码。它们共同起作用,允许识别正在寻址的蛋白质,使其展开(蛋白质必须保持在非折叠的状态才能穿过膜),然后导入,随后切割寻址肽,最终将蛋白精确定位到其所在功能区[19]。
这种在细胞器中发挥功能的细胞核编码基因,究竟是如何起源的?实际上存在两种可能(图7)[16]。有时,原始核基因取代了细胞器基因:对应的基因产物能够精确定位到细胞器中发挥功能。然而,如果细胞器中已经存在了一个编码相同功能的基因,那么这种定位转移可能导致蛋白质的冗余。由于这种冗余,细胞器基因可能逐渐丢失而不损害其功能(图7a)[16]。
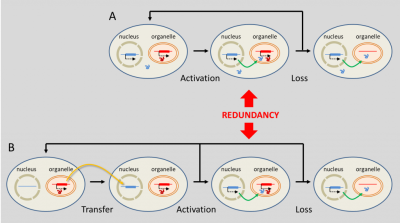
(A)“真正的”细胞核起源的基因取代了细胞器基因;(B)细胞器向细胞核方向的基因转移。[来源:根据瑟洛斯(Selosse)等人(2001年)参考文献[16]]。(图7 nucleus 细胞核;organelle 细胞器;Activation 活化;Loss 丢失;Redundancy 冗余;Transfer 转移)
DNA片段从细胞器转移到细胞核并不罕见:某些植物的基因组中插入了大片段的细胞器DNA。这些基因可以被激活:拟南芥中近10%的核基因是从质体转移而来的,其次是复制子[20]。目前尚不清楚内共生体的DNA是如何融合到宿主基因组中的,但人们认为,这可能发生在受损或衰老的细胞器降解过程中,DNA片段意外地释放到宿主细胞质,然后随机融合到宿主核DNA中。
细胞器的细胞质基因组处于各种选择压力下,其中一些选择力促进它们的退化(例如某些基因的共表达需要),另一些选择力有利于特定基因在细胞器基因组中保留。细胞器小基因组选择可能就是这种情况,它加速了细胞器的繁殖,允许更好地传递遗传信息给子代细胞:例如,它选择将基因转移到细胞核。因此,它积累了来自不同谱系的遗传潜力,这些谱系与它在细胞中共存[16]。因此,虽然内共生减少了内共生体的基因组,但它滋养了宿主细胞核的基因组,促进了核基因组的遗传多样化,并推动了更紧密的内共生关系形成。因此,通过多次内共生嵌套以及宿主细胞核基因嵌合,内共生体逐渐构成了目前的进化谱系。
最后,内共生体的跨代垂直转移对于内共生体的持续存在至关重要。质体必须在宿主细胞分裂前分裂,并且必须均匀分布在两个子细胞中。如果分裂得太快,它们可能比宿主细胞占优势。相反,分裂得太慢则可能导致内共生体消失。在这种情况下,细胞分裂和共生体分裂的协调进行是内共生成功的关键因素。虽然参与叶绿体分裂的大多数蛋白质来自蓝细菌的细胞分裂机制,但有些蛋白质显然是真核来源的(这种蛋白都在细胞核中编码):这是宿主控制叶绿体分裂的一种方式。
5.共生驱动进化吗?
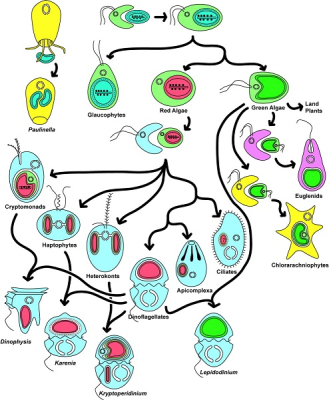
由此产生了各种各样的生物,如红藻和绿藻、陆生植物、顶复门生物(如疟原虫、弓形虫,可导致疟疾)或甲藻(一种海洋浮游生物,在初级海洋生产中尤为重要)。[来源:转载自基林(Keeling)等人[12]。 ©2016 美国植物学杂志版权所有](图8 Paulinella 阿米巴原虫;Glaucophytes 灰胞藻;Red Algae 红藻;Green Algae 绿藻;Land Plants 陆地植物;Euglena 裸藻;Chlorarachniophytes 海洋阿米巴鞭毛原虫;Cryptomonads 隐藻;Haptophytes 定鞭金藻;Heterokonts 不等鞭毛藻;Dinoflagellates 甲藻;Apicomplexa 顶复门生物;Ciliates 纤毛虫;Dinophysis 鳍藻;Karenia 凯伦藻;Kryptoperidinium 隐多甲藻;Lepidodinium 鳞甲藻)
综上所述,这种多样化的共生关系导致了真核细胞的形成[1,9],是真核生物进化过程中多样性发展的基础。内共生是新的进化谱系形成的基础。图8展示了内共生生物的极端多样性。然而,事情远比想象的复杂:这种共生进化过程可能会持续重复!今天,一些独特的单细胞藻类,如隐藻【一种单细胞生物,多数能进行光合作用。它们的叶绿体由四个膜包围,表明它与能够进行光合作用的真核生物进行了内共生。隐藻存在于许多环境中,特别是水生环境(海洋环境、淡水、湿地孔隙水)。一些物种已经成为后生动物的肠道寄生虫。有些是甲藻的内共生体。】和异形藻(图8),由二次内共生质体进一步融合到甲藻细胞质内形成(这种情况下,甲藻失去了原有的质体)。它们的质体含有四层膜,属于二次内共生质体:这里发生了三次连续的内共生!
共生不仅仅是一种生物学现象,它还是推动生命世界进化的强大动力之一。它能迅速创造出嵌合生物,可以产生新的细胞系。它拉近了共生伙伴间的距离,促进了大规模的基因转移,创造了嵌合基因组:这种核基因组包含真核基因,同时也包含细菌基因(来源于线粒体,甚至与之相邻生物的质体)。这些多样的共生事件可以解释生命进化史上的重大飞跃(不连续的进化事件),它们创造了强大的生命,塑造了当前的生物多样性。
因此,从达尔文进化观点来看,一个物种可能会产生两个物种(即所有物种来自于一个共同祖先),但内共生机制提醒我们,有时两个物种(自由生活的、独立可区分的)也可能合并成一个新的物种。人类可以被看作是一个高度融合的共生群落,由真核细胞质和线粒体(起源于原核生物)组成,同时也包括古菌和细菌(例如肠道微生物)。
参考资料和说明
[1] Lang T. et al (2000) Autophagy and the cvt pathway both depend on AUT9. J Bacteriol 182, 2125-2133.
[2] Chatton E. (1938) Titres et travaux scientifiques (1906-1937). Sette, Sottano, Italy. L’histoire des conditions dans lesquelles Chatton a établi le concept de procaryote et eucaryote est décrite par Sapp J. (2005) The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology, Microbiol Mol Biol Rev. 69, 292–305.
[3] Mereschkowsky C. 1905 Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche. Centralbl. 25, 593-604; translated by Martin W, Kowallik K. (1999) Annotated English translation of Mereschkowsky’s 1905 paper ‘Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche‘. Eur. J. Phycol. 34, 287–295.
[4] Portier P. (1918) Les Symbiotes. Masson (ed.), Paris. (in french)
[5] Martin W.F., Garg S. & Zimorski V. (2015) Endosymbiotic theories for eukaryote origin. Phil. Trans. R. Soc. B370, 20140330.
[6] Selosse M.A. (2012). Gloire et disgrâce de la théorie endosymbiotique. La Recherche 468: 92-94. (in french)
[7] Archibald J.M. (2014) One plus one equals one: symbiosis and the evolution of complex life. Oxford, UK: Oxford University Press.
[8] McFadden G.I. (2014) Origin and Evolution of Plastids and Photosynthesis in Eukaryotes, Cold Spring Harb.Perspect. Biol. 6, a016105
[9] Martin W. & Müller M. (1998) The hydrogen hypothesis for the first eukaryote. Nature 392, 37-41.
[10] Ettema T.J.G. (2016) Mitochondria in the second act. Nature 531, 39-40doi:10.1038/nature16876
[11] Archibald J.M. & Keeling P.J. (2002) Recycled plastids: a “green movement” in eukaryotic evolution. Trends Genetics 18, 577-584.
[12] Keeling P.J. (2004) Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 91, 1481-1493.
[13] Douce R., Block M.A., Dorne A.J., Joyard J. (1984) The plastid envelope membranes: their structure, composition, and role in chloroplast biogenesis. Subcell. Biochem. 10, 1-84, Springer US (Ed.)
[14] Selosse M.A. & Loiseaux-de Goër S. (1997) La Saga de l’endosymbiose, La Recherche 296, 36 (in french)
[15] Embley T.M. & Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440, 623-630
[16] Lefèvre T., Renaud F., Selosse M.-A. & Thomas F. (2010). Évolution des interactions entre espèces, in F. Thomas, T. Lefèvre & M. Raymond (ed.), Biologie évolutive, p. 530-613. De Boeck, Paris. (in french)
[17] Keeling P.J. (2010) The endosymbiotic origin, diversification and fate of plastids. Phil. Trans. R. Soc. B 365, 729-748
[18] Douglas S. et al (2001) The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091-1096.
[19] Selosse M.A., Albert B. & Godelle B. (2001) Small is successful: selection for reducing organelle’s genome size favours gene transfer to the nucleus. Trends Ecol Evol 16, 135-141.
[20] Jarvis P. (2004) Organellar Proteomics: Chloroplasts in the Spotlight. Current Biology 14, R317-9. http://www.cell.com/current-biology/references/S0960-9822%2804%2900231-3
环境百科全书由环境和能源百科全书协会出版 (www.a3e.fr),该协会与格勒诺布尔阿尔卑斯大学和格勒诺布尔INP有合同关系,并由法国科学院赞助。
引用这篇文章: SELOSSE Marc-André, JOYARD Jacques (2024年3月12日), 共生与进化:真核细胞的起源, 环境百科全书,咨询于 2025年1月21日 [在线ISSN 2555-0950]网址: https://www.encyclopedie-environnement.org/zh/vivant-zh/symbiosis-and-evolution-origin-eukaryotic-cell/.
环境百科全书中的文章是根据知识共享BY-NC-SA许可条款提供的,该许可授权复制的条件是:引用来源,不作商业使用,共享相同的初始条件,并且在每次重复使用或分发时复制知识共享BY-NC-SA许可声明。